Model evaluation
CSI 4106 - Fall 2024
Version: Oct 11, 2024 10:39
Preamble
Quote of Day
Learning objectives
- Clarify the concepts of underfitting and overfitting in machine learning.
- Describe the primary metrics used to evaluate model performance.
- Contrast micro- and macro-averaged performance metrics.
Model fitting
Model fitting
During our class discussions, we have touched upon the concepts of underfitting and overfitting. To delve deeper into these topics, let’s examine them in the context of polynomial regression.
Generating a nonlinear dataset
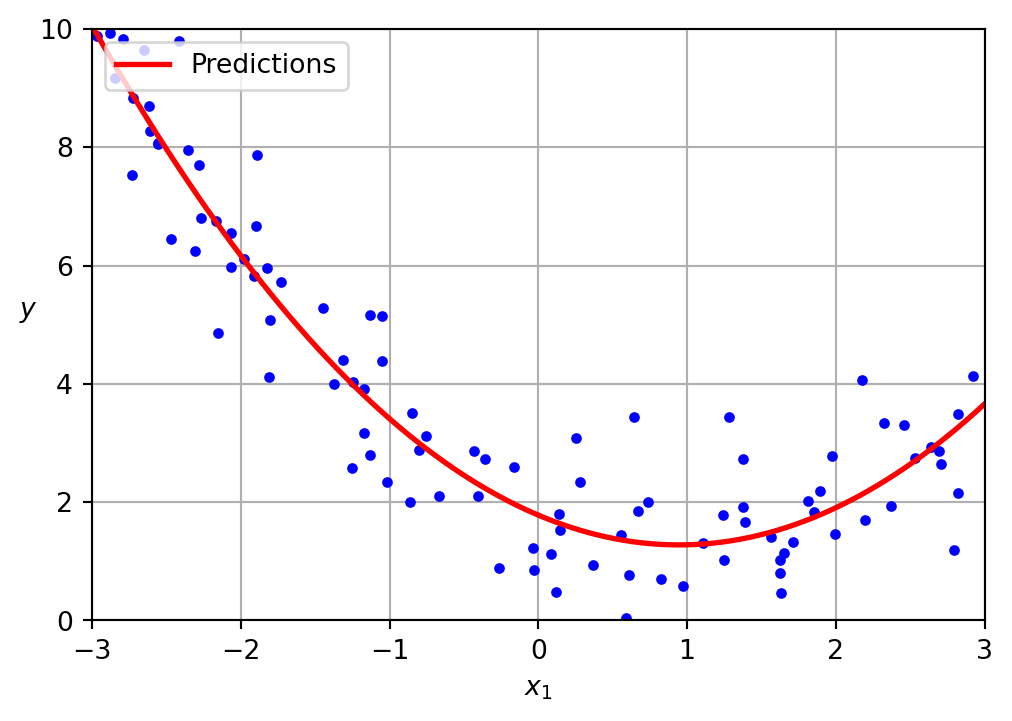
Linear regression
A linear model inadequately represents this dataset
Definition
Feature engineering is the process of creating, transforming, and selecting variables (attributes) from raw data to improve the performance of machine learning models.
PolynomialFeatures
Generate a new feature matrix consisting of all polynomial combinations of the features with degree less than or equal to the specified degree. For example, if an input sample is two dimensional and of the form \([a, b]\), the degree-2 polynomial features are \([1, a, b, a^2, ab, b^2]\).
PolynomialFeatures
Given two features \(a\) and \(b\), PolynomialFeatures with degree=3 would add \(a^2\), \(a^3\), \(b^2\), \(b^3\), as well as, \(ab\), \(a^2b\), \(ab^2\)!
Warning
PolynomialFeatures(degree=d) adds \(\frac{(D+d)!}{d!D!}\) features, where \(D\) is the original number of features.
Polynomial regression
LinearRegression on PolynomialFeatures
Polynomial regression
The data was generated according to the following equation, with the inclusion of Gaussian noise.
\[ y = 0.5 x^2 + 1.0 x + 2.0 \]
Presented below is the learned model.
\[ \hat{y} = 0.56 x^2 + (-1.06) x + 1.78 \]
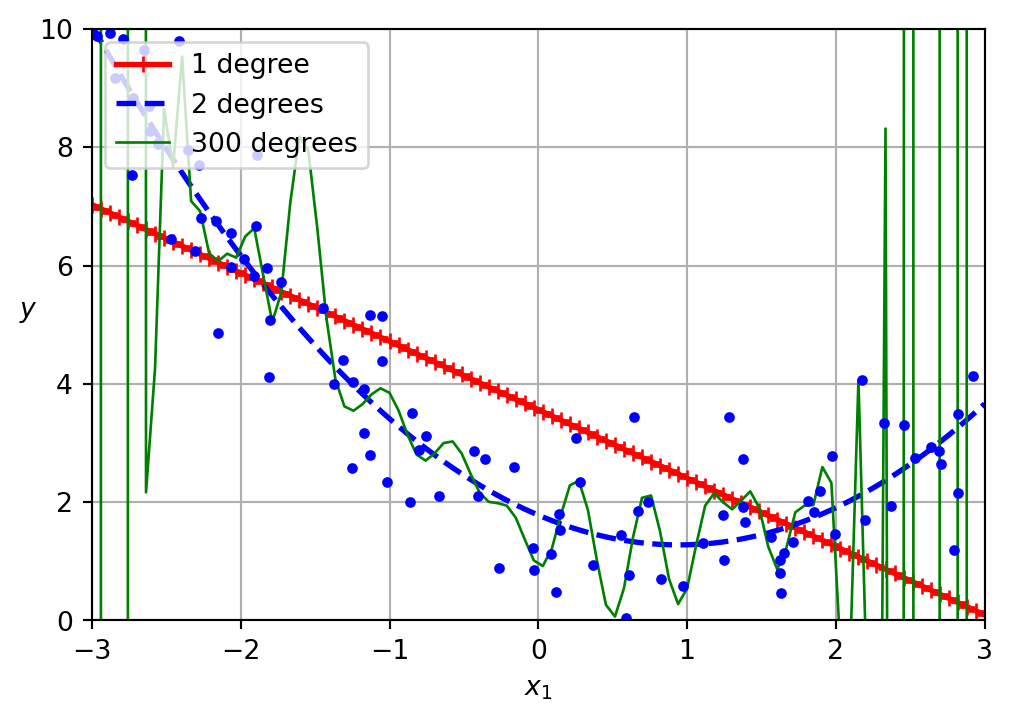
Overfitting and underfitting
A low loss value on the training set does not necessarily indicate a “better” model.
Under- and over- fitting
- Underfitting:
- Your model is too simple (here, linear).
- Uninformative features.
- Poor performance on both training and test data.
- Overfitting:
- Your model is too complex (tall decision tree, deep and wide neural networks, etc.).
- Too many features given the number of examples available.
- Excellent performance on the training set, but poor performance on the test set.
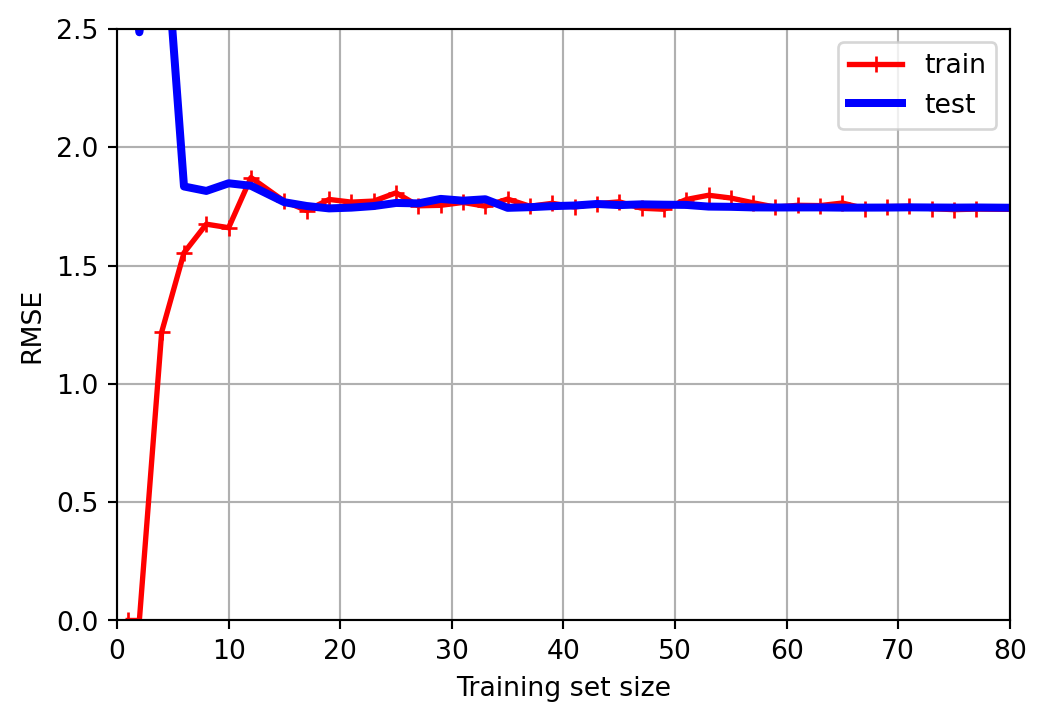
Learning curves
- One way to assess our models is to visualize the learning curves:
- A learning curve shows the performance of our model, here using RMSE, on both the training set and the test set.
- Multiple measurements are obtained by repeatedly training the model on larger and larger subsets of the data.
Learning curve – underfitting
Poor performance on both training and test data.
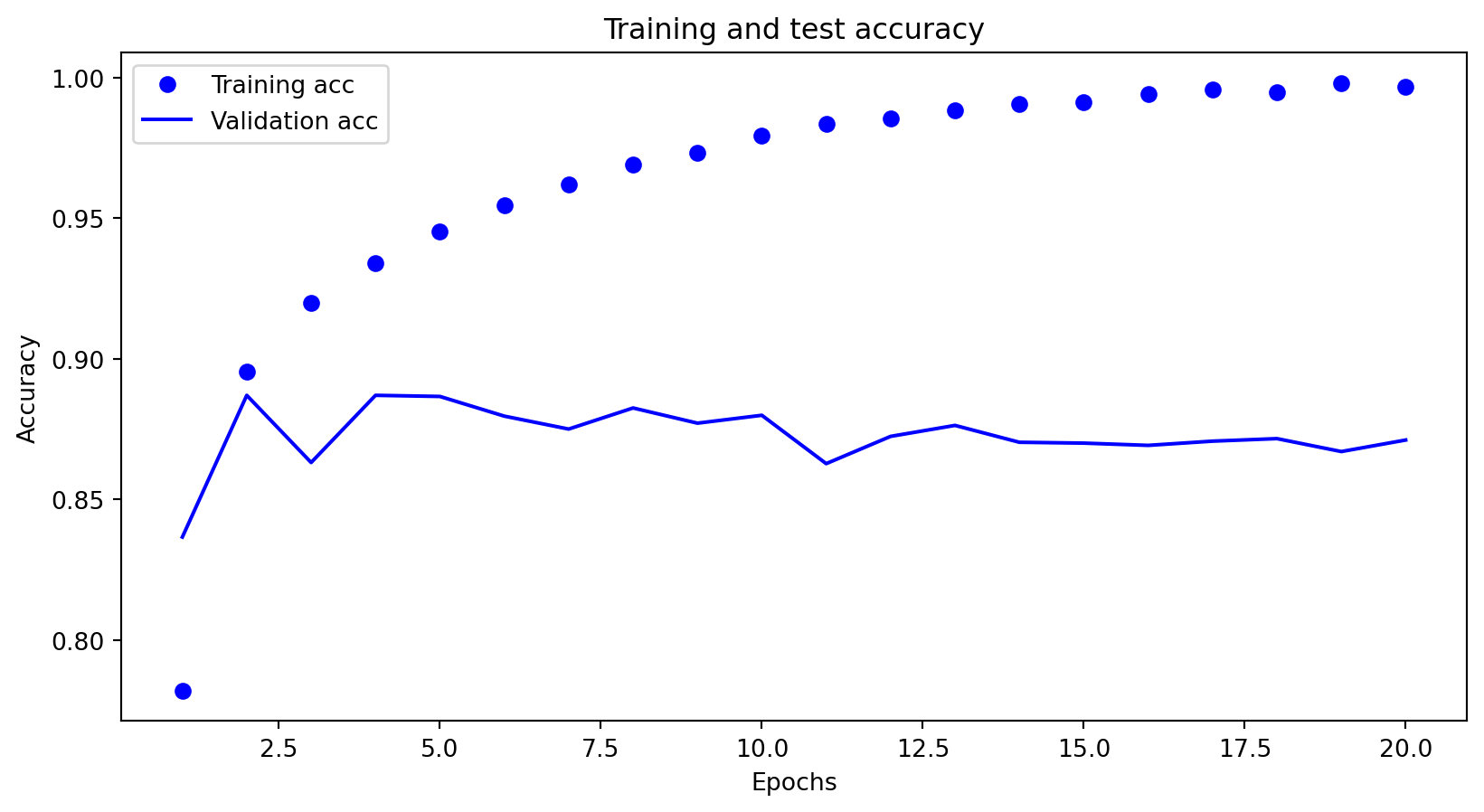
Learning curve – overfitting
Excellent performance on the training set, but poor performance on the test set.
Overfitting - deep nets - loss
Overfitting - deep nets - accuracy
Bias/Variance Tradeoff
- Bias:
- Error from overly simplistic models
- High bias can lead to underfitting
- Variance:
- Error from overly complex models
- Sensitivity to fluctuations in the training data
- High variance can lead to overfitting
- Tradeoff:
- Aim for a model that generalizes well to new data
- Methods: cross-validation, regularization, ensemble learning
Related videos
Other videos include:
Performance metrics
Confusion matrix
| Positive (Predicted) | Negative (Predicted) | |
|---|---|---|
| Positive (Actual) | True positive (TP) | False negative (FN) |
| Negative (Actual) | False positive (FP) | True negative (TN) |
sklearn.metrics.confusion_matrix
Perfect prediction
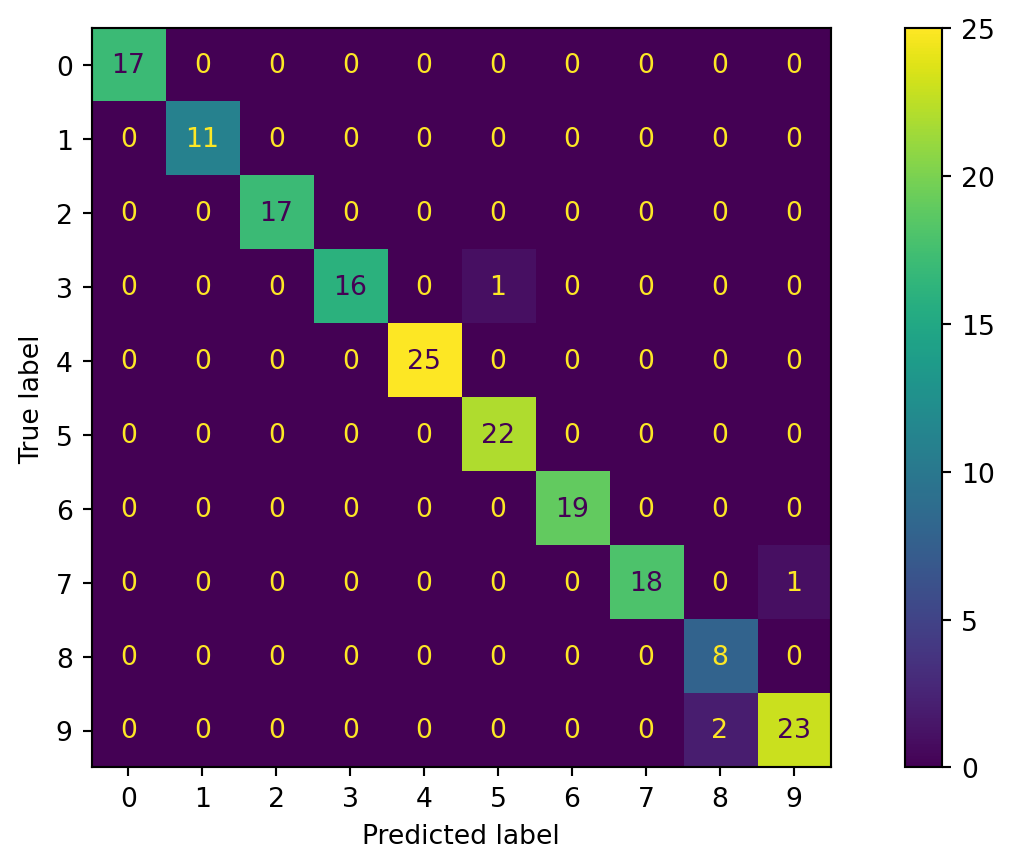
Confusion matrix - multiple classes
Source code
import numpy as np
np.random.seed(42)
from sklearn.datasets import load_digits
digits = load_digits()
X = digits.data
y = digits.target
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
from sklearn.linear_model import LogisticRegression
from sklearn.multiclass import OneVsRestClassifier
clf = OneVsRestClassifier(LogisticRegression())
clf = clf.fit(X_train, y_train)
import matplotlib.pyplot as plt
from sklearn.metrics import ConfusionMatrixDisplay
X_test = scaler.transform(X_test)
y_pred = clf.predict(X_test)
ConfusionMatrixDisplay.from_predictions(y_test, y_pred)
plt.show()Visualizing errors
Confusion matrix - multiple classes
Accuracy
How accurate is this result?
\[ \mathrm{accuracy} = \frac{\mathrm{TP}+\mathrm{TN}}{\mathrm{TP}+\mathrm{TN}+\mathrm{FP}+\mathrm{FN}} = \frac{\mathrm{TP}+\mathrm{TN}}{\mathrm{N}} \]
Accuracy
Accuracy can be misleading
Precision
AKA, positive predictive value (PPV).
\[ \mathrm{precision} = \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FP}} \]
Precision alone is not enough
Recall
AKA sensitivity or true positive rate (TPR) \[ \mathrm{recall} = \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FN}} \]
F\(_1\) score
\[ \begin{align*} F_1~\mathrm{score} &= \frac{2}{\frac{1}{\mathrm{precision}}+\frac{1}{\mathrm{recall}}} = 2 \times \frac{\mathrm{precision}\times\mathrm{recall}}{\mathrm{precision}+\mathrm{recall}} \\ &= \frac{\mathrm{TP}}{\mathrm{FP}+\frac{\mathrm{FN}+\mathrm{FP}}{2}} \end{align*} \]
Micro Performance Metrics
Micro performance metrics aggregate the contributions of all classes to compute the average performance metric, such as precision, recall, or F1 score. This approach treats each individual prediction equally, providing a balanced evaluation by emphasizing the performance on frequent classes.
Macro Performance Metrics
Macro performance metrics compute the performance metric independently for each class and then average these metrics. This approach treats each class equally, regardless of its frequency, providing an evaluation that equally considers performance across both frequent and infrequent classes.
Micro/macro metrics
from sklearn.metrics import ConfusionMatrixDisplay
# Sample data
y_true = ['Cat'] * 42 + ['Dog'] * 7 + ['Fox'] * 11
y_pred = ['Cat'] * 39 + ['Dog'] * 1 + ['Fox'] * 2 + \
['Cat'] * 4 + ['Dog'] * 3 + ['Fox'] * 0 + \
['Cat'] * 5 + ['Dog'] * 1 + ['Fox'] * 5
ConfusionMatrixDisplay.from_predictions(y_true, y_pred)Micro/macro precision
from sklearn.metrics import classification_report, precision_score
print(classification_report(y_true, y_pred), "\n")
print("Micro precision: {:.2f}".format(precision_score(y_true, y_pred, average='micro')))
print("Macro precision: {:.2f}".format(precision_score(y_true, y_pred, average='macro'))) precision recall f1-score support
Cat 0.81 0.93 0.87 42
Dog 0.60 0.43 0.50 7
Fox 0.71 0.45 0.56 11
accuracy 0.78 60
macro avg 0.71 0.60 0.64 60
weighted avg 0.77 0.78 0.77 60
Micro precision: 0.78
Macro precision: 0.71Macro-average precision is calculated as the mean of the precision scores for each class: \(\frac{0.81 + 0.60 + 0.71}{3} = 0.71\).
Whereas, the micro-average precision is calculated using the formala, \(\frac{TP}{TP+FP}\) and the data from the entire confusion matrix \(\frac{39+3+5}{39+3+5+9+2+2} = \frac{47}{60} = 0.78\)
Micro/macro recall
precision recall f1-score support
Cat 0.81 0.93 0.87 42
Dog 0.60 0.43 0.50 7
Fox 0.71 0.45 0.56 11
accuracy 0.78 60
macro avg 0.71 0.60 0.64 60
weighted avg 0.77 0.78 0.77 60
Micro recall: 0.78
Macro recall: 0.60Macro-average recall is calculated as the mean of the recall scores for each class: \(\frac{0.93 + 0.43 + 0.45}{3} = 0.60\).
Whereas, the micro-average recall is calculated using the formala, \(\frac{TP}{TP+FN}\) and the data from the entire confusion matrix \(\frac{39+3+5}{39+3+5+3+4+6} = \frac{39}{60} = 0.78\)
Micro/macro metrics (medical data)
Consider a medical dataset, such as those involving diagnostic tests or imaging, comprising 990 normal samples and 10 abnormal (tumor) samples. This represents the ground truth.
Micro/macro metrics (medical data)
precision recall f1-score support
Normal 1.00 0.99 1.00 990
Tumour 0.55 0.60 0.57 10
accuracy 0.99 1000
macro avg 0.77 0.80 0.78 1000
weighted avg 0.99 0.99 0.99 1000
Micro precision: 0.99
Macro precision: 0.77
Micro recall: 0.99
Macro recall: 0.80Hand-written digits (revisited)
Loading the dataset
Plotting the first five examples
These images have dimensions of ( 28 ) pixels.
Creating a binary classification task
SGDClassifier
Performance
from sklearn.metrics import accuracy_score
y_pred = clf.predict(X_test)
accuracy_score(y_test, y_pred)0.9572857142857143Wow!
Not so fast
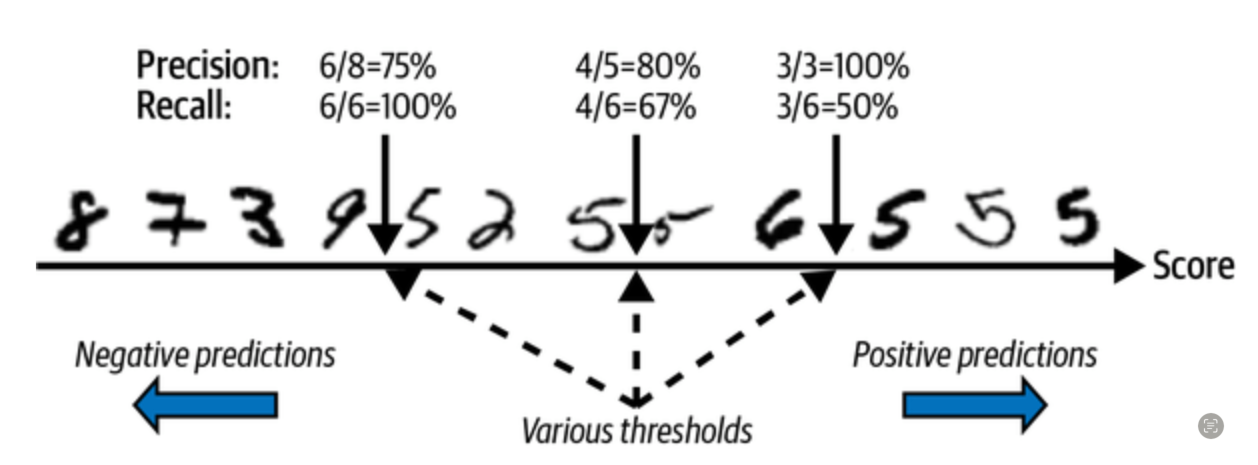
Precision-recall trade-off
Precision-recall trade-off
Precision/Recall curve
ROC curve
Receiver Operating Characteristics (ROC) curve
- True positive rate (TPR) against false positive rate (FPR)
- An ideal classifier has TPR close to 1.0 and FPR close to 0.0
- \(\mathrm{TPR} = \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FN}}\) (recall, sensitivity)
- TPR approaches one when the number of false negative predictions is low
- \(\mathrm{FPR} = \frac{\mathrm{FP}}{\mathrm{FP}+\mathrm{TN}}\) (aka~[1-specificity])
- FPR approaches zero when the number of false positive is low
ROC curve
AUC/ROC
The 7 steps of machine learning
Prologue
Further reading
Next lecture
- We will examine cross-validation and hyperparameter tuning.
References
Marcel Turcotte
School of Electrical Engineering and Computer Science (EECS)
University of Ottawa