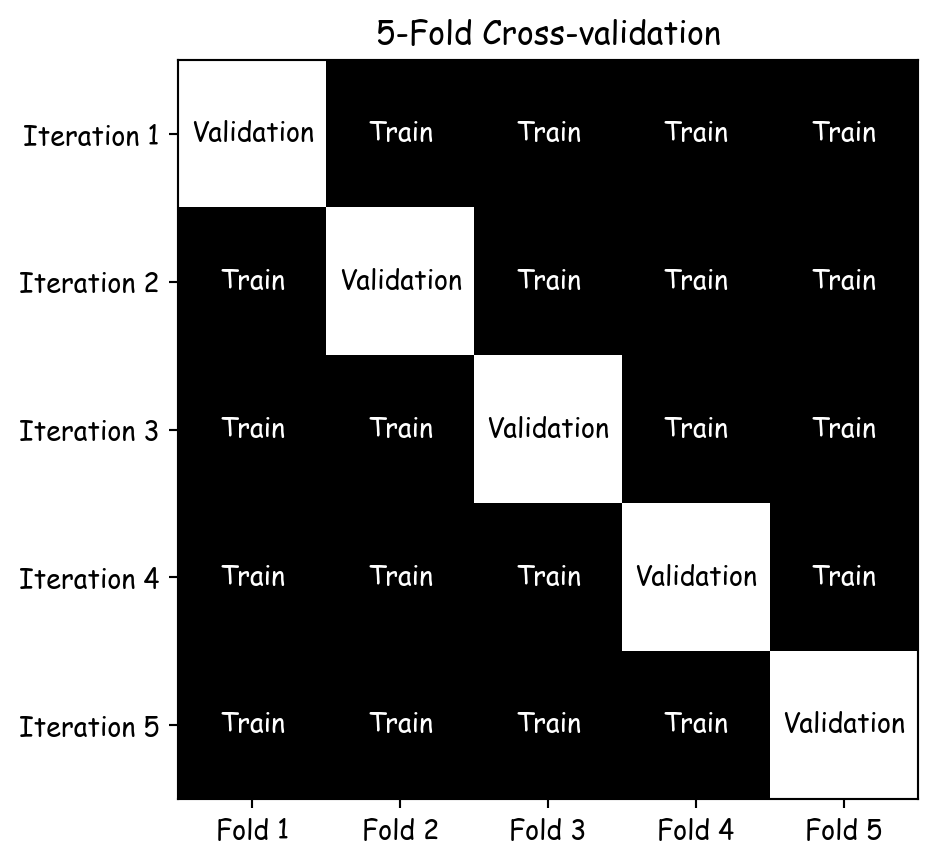
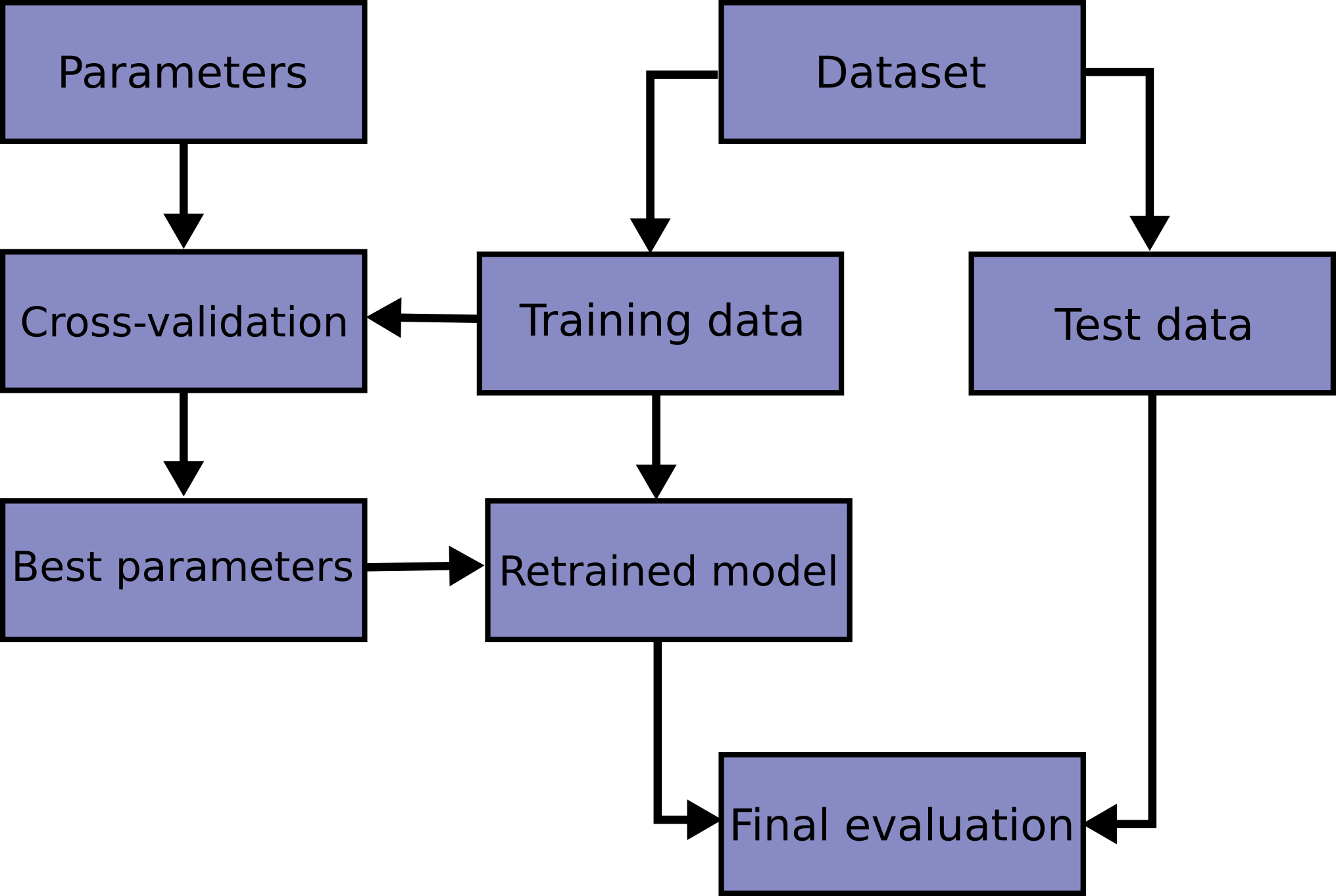
import numpy as np
np.random.seed(42)
from sklearn.datasets import fetch_openml
diabetes = fetch_openml(name='diabetes', version=1)
print(diabetes.DESCR)Author: Vincent Sigillito
Source: Obtained from UCI
Please cite: UCI citation policy
Title: Pima Indians Diabetes Database
Sources:
- Original owners: National Institute of Diabetes and Digestive and Kidney Diseases
- Donor of database: Vincent Sigillito ([email protected]) Research Center, RMI Group Leader Applied Physics Laboratory The Johns Hopkins University Johns Hopkins Road Laurel, MD 20707 (301) 953-6231
- Date received: 9 May 1990
Past Usage:
Smith,J.W., Everhart,J.E., Dickson,W.C., Knowler,W.C., & Johannes,R.S. (1988). Using the ADAP learning algorithm to forecast the onset of diabetes mellitus. In {it Proceedings of the Symposium on Computer Applications and Medical Care} (pp. 261–265). IEEE Computer Society Press.
The diagnostic, binary-valued variable investigated is whether the patient shows signs of diabetes according to World Health Organization criteria (i.e., if the 2 hour post-load plasma glucose was at least 200 mg/dl at any survey examination or if found during routine medical care). The population lives near Phoenix, Arizona, USA.
Results: Their ADAP algorithm makes a real-valued prediction between 0 and 1. This was transformed into a binary decision using a cutoff of 0.448. Using 576 training instances, the sensitivity and specificity of their algorithm was 76% on the remaining 192 instances.
Relevant Information: Several constraints were placed on the selection of these instances from a larger database. In particular, all patients here are females at least 21 years old of Pima Indian heritage. ADAP is an adaptive learning routine that generates and executes digital analogs of perceptron-like devices. It is a unique algorithm; see the paper for details.
Number of Instances: 768
Number of Attributes: 8 plus class
For Each Attribute: (all numeric-valued)
- Number of times pregnant
- Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- Diastolic blood pressure (mm Hg)
- Triceps skin fold thickness (mm)
- 2-Hour serum insulin (mu U/ml)
- Body mass index (weight in kg/(height in m)^2)
- Diabetes pedigree function
- Age (years)
- Class variable (0 or 1)
Missing Attribute Values: None
Class Distribution: (class value 1 is interpreted as “tested positive for diabetes”)
Class Value Number of instances 0 500 1 268
Brief statistical analysis:
Attribute number: Mean: Standard Deviation:
3.8 3.4120.9 32.069.1 19.420.5 16.079.8 115.232.0 7.90.5 0.333.2 11.8
Relabeled values in attribute ‘class’ From: 0 To: tested_negative
From: 1 To: tested_positive
Downloaded from openml.org.